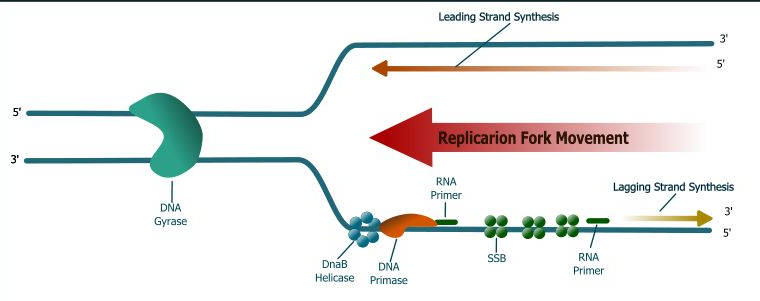
DNA elongation is a complex process involving cooperative interactions among many different proteins. These proteins synthesize two new DNA chains at an astonishing rate. Tania Baker and Stephen Bell provide the following insightful analogy to illustrate the astonishing job that the E.coli replication enzymes perform while synthesizing DNA. They begin by changing the scale so that the DNA duplex is 1 m in diameter. At this scale, the replication fork would move at about 600 km/h (375 mph) and the replication machinery would be about the size of a FedEx® delivery truck.
Baker and Bell's analogy underscores the enormous complexity and incredible speed of the DNA replication process. DNA polymefase Ⅲ holoenzyme, the bacterial counterpart of the FedEx delivery truck, acts as the replication machine. It extends the leading strand by continuous nucleotide attachment to the 3'-hydroxy terminus, while helping to form the lagging strand by extending the primer until the resulting Okazaki fragment reaches the 5'-end of the adjacent fragment. Although DNA polymerase Ⅲ holoenzyme is a remarkable replication machine, it does not act alone. Several additional proteins assist in DNA synthesis at the replication fork. These events, which are summarized, are as follows:
1. DnaB, a homohexameric helicase, use energy supplied by ATP hydrolysis to unwind double-stranded DNA as the helicase moves 5'→3'along the lagging template strand. Unwinding at the replication fork leads to tighter winding ahead of the replication fork because the bacterial chromosome is a double-stranded circular DNA molecule. Helicase also activates RNA primase (see below) to make the RNA primers.
2. DNA gyrase and topoisomerase I act cooperatively to relieve the resulting torsional strain, allowing the replication fork to continue moving along the bacterial chromosome.
3. Single-stranded binding protein (SSB) coats the lagging strand template exposed by helicase action so that it is protected from nuclease attack and cannot form intrastrand base pairs.
4. Primase (DnaG protein) catalyzes the formation of short RNA oligomers that then serve as primers for the replication machine. Primase needs to act just once to initiate leading strand synthesis because chain extension is continuous, but must act repeatedly during lagging strand synthesis to initiate Okazaki fragment synthesis once in every 500 to 2000 nucleotides.
5. Ribonuclease H (RNase H) removes all of the ribonucleotides at the 5'-terminus of the Okazaki fragment except for the ribonucleotide that is linked to the DNA end.
6. DNA polymerase 1 catalyzes nick translation. Its polymerase activity extends the 3'-end of the newly formed Okazaki fragment as its 5'→3' exonuclease removes the ribonucleotide from the 5'-terminus of the neighboring Okazaki fragment.
7. DNA ligase joins adjacent Okazaki fragments to form the lagging strand.