Posted by star
on 2019-06-03 23:34:26
Hits:255
The antibiotic resistance among pathogenic bacteria is one of the biggest problems in public health. Bacterial geneticists at the University of Lyon have found that E. coli can synthesize drug-resistant proteins even in the presence of antibiotics. A well-preserved membrane pump removes antibiotics from the cells, allowing long enough time for the cells to receive DNA from neighboring cells that encode a resistance protein.
The researchers used the normal antibiotic resistance gene of E. coli as the research object, and carefully observed how the bacteria encode the TetA protein by DNA (the TetA protein is a pump that makes cells resistant to tetracycline by shunting out tetracycline). They found that after the plasmid DNA reached the non-resistant cells for a period of time, red fluorescence appeared on the receptor cell membrane, indicating that the TetA protein was translated and the cells were resistant to tetracycline.
The researchers then exposed the cells to high concentrations of tetracycline and placed them under the microscope again. They observed that plasmid DNA reached new non-resistant cells. This is expected because tetracycline does not hinder this process. However, it prevents protein production.
But the researchers found red fluorescence in some of the new recipient cells, which previously had no TetA protein. Obviously, despite exposure to tetracycline, they are still able to synthesize proteins, including TetA.
The researchers found that the AcrAB-TolC pump can prolong the survival of bacteria by keeping the concentration of antibiotics low enough to allow cells to synthesize resistance proteins encoded in plasmid DNA. In this case, it can product TetA protein and then separate more tetracycline from the cells. Ultimately, bacteria can become resistant under the influence of antibiotics.
This finding has broad relevance because AcrAB-TolC is highly conserved in bacteria and because its mechanism is not limited to tetra......
Posted by star
on 2019-06-03 23:22:17
Hits:284
Given the central role of the ER in the synthesis of proteins for the entire exocytic and endocytic pathways, it is not surprising that many inherited diseases are a direct consequence of proteins failing to pass ER quality control. Many metabolic disorders, including some lysosomal storage diseases, are a direct consequence of key enzymes failing to be exported from the ER. The most common form of cystic fibrosis is due to the inability of cells to export a mutant form of the cystic fibrosis transmembrane regulator to the cell surface, where it would normally function as a chloride channel for the respiratory system and pancreas. Similarly, the inability of the liver to secrete mutated forms of a1- antitrypsin predisposes to the lung dis-ease emphysema. Normally, a1-antitrypsinprotects tissues by inhibiting extracellular proteases such as elastase, which is produced by neutrophils. Mutations prevent a1-antitrypsinfolding in the ER, resulting in its degradation. The resulting deficiency in a1-antitrypsin circulating in the blood allows elastase to destroy lung tissue, leading to emphysema. In severe cases, mutant forms of the protein not only fail to be exported from the ER but also elude degradation path-ways, accumulating as insoluble aggregates that induce stress responses and liver failure.
In some conditions, the ER uses the unfolded protein response to compensate, in part, for mutations in cargo proteins. In congenital hypothyroidism, mutant thyroglobulin is not exported efficiently from the ER. Excess protein accumulates as in soluble aggregates in the ER. Feedback pathways trigger massive proliferation of ER in an attempt to produce normal levels of circulating hormone. Similarly, in mild forms of osteogenesis imperfecta, osteoblasts assemble and secrete defective procollagen chains for bone synthesis, even though the resulting bone tissue is weak. 'The alternative, complete loss of procollagen by retention and degradation of the mutant procollagen i......
Posted by star
on 2019-06-02 19:44:04
Hits:281
The human MT1 and MT2 melatonin receptors1,2 are G-proteincoupled receptors (GPCRs) that help to regulate circadian rhythm and sleep patterns. Drug development efforts have targeted both receptors for the treatment of insomnia, circadian rhythm and mood disorders, and cancer, and MT2 has also been implicated in type 2 diabetes.
Melatonin is produced by the pineal gland in the center of the brain. Humans respond naturally to changes in sunlight through the pineal gland near the hypothalamus. As night falls, the glands produce more melatonin and then bind to the MT1 and MT2 receptors of the cells. Before dawn, the gland will lower the level of melatonin, which indicates that it is time to wake up.
Recently, a team of international scientists has developed a new method for treating sleep disorders based on two new 3D models of melatonin receptors.
The researchers created a 3D map of two melatonin receptors MT1 and MT2. They hope to use this information to design drug molecules that bind to melatonin receptors and monitor their potential effects. This will treat a variety of diseases in a more targeted manner, including diabetes, cancer and sleep disorders.
"By comparing the three-dimensional structure of the MT1 and MT2 receptors, we can better distinguish the unique structural differences between the two receptors and their role in the circadian clock. With this knowledge, the design is only one or Another receptor binds, but the drug-like molecules that do not simultaneously bind to the two receptors become easier. This selective binding is important because it reduces unwanted side effects." the researcher said:
The structure of these two receptors was obtained by using a laser called the LCLS linear continuous accelerator source in the SLAC National Accelerator Laboratory. This laser uses X-rays to capture stop-work photos of receptor atoms and molecules in motion.
![]()
Posted by star
on 2019-06-02 19:30:39
Hits:256
Given a mutation in a gene of interest, two genetic tests are used to search for partners: (1) identification of a second mutation that ameliorates the effects of the primary mutation and (2) identification of a second mutation that makes the phenotype more severe, often lethal. A specialized class of enhancer mutations, called synthetic lethal mutations, is particularly useful in the analysis of genetic pathways in yeast. In this case, mutations in two genes in the same pathway, if present in the same cell, even as heterozygotes, cannot be tolerated, so the cell dies. It is thought that each mutation lowers the level of production of some critical factor just a bit and that the combination of the two effectively means that the output of the pathway is insuffcient for survival. These tests can be made with existing collections of mutations by genetically crossing mutant organisms. Alternatively, one can seek new mutations created by a second round of mutagenesis. The results depend on the architecture of the particular pathway. If the products of the genes in question operate in a sequence, analysis of single and double mutants can often reveal their order in the pathway. For essential genes in haploid organisms, a conditional allele of the primary mutation simplifies the experiment. Synthetic interactions may also be discovered by overproduction of wild-type genes on a plasmid. Caution is required in interpreting suppressor and enhancer mutations, given the complexity of cellular systems and the possibility of unanticipated consequences of the mutations.

Another approach to find protein partners is called a two-hybrid assay. This assay depends on the observation that some activators of transcription have two modular domains with discrete functions: One domain binds target sites on DNA, and ......
Posted by star
on 2019-05-30 23:35:04
Hits:296
The researchers found that chronic gastritis and helicobacter pylori, scientists subsequently carried out a large study of helicobacter pylori, the research results show that in addition to cause gastritis, h. pylori is also the key factors that induce gastric ulcer and gastric cancer, the researchers clearly illustrates the associated bacteria and a variety of stomach disease, but how is it that h. pylori induced gastric cancer, the scientific community there are a lot of controversy.
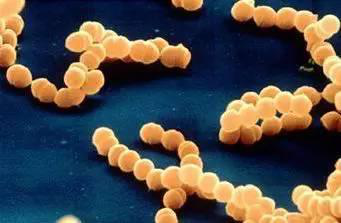
Scientists from a Japanese research institute have revealed the molecular mechanism by which helicobacter pylori inflammation promotes the proliferation of gastric epithelial stem cells, which then trigger tumours in the stomach.
Researcher Dr Kamil Kranc said: we have previously shown that TNF- alpha, which promotes inflammation in the body, may promote gastric tumours by activating the function of NOXO1, but it is not clear how NOXO1 induces tumours in the stomach.NOXO1 is a component of NOX1 complex, which can produce tissue damaging molecular reactive oxygen species (ROS), and the oxidative stress reaction induced by ROS will lead to DNA mutation in gastric cells, which will lead to tumor formation, and the inflammation caused by helicobacter pylori infection will also produce ROS, which will increase the oxidative stress reaction of stomach.
Researchers found that excessive inflammation promotes NOX1 compound protein produced by the reaction of the NF-κB signal, the NF-κB is a kind of regulatory protein, it can be open to withstand pressure or bacterial infection of the expression of genes, is also important role in the process of inflammatory reaction, more importantly, however, the researchers found that NOX1 / ROS can promote gastric epithelial stem cell......
